New preprint on mutational spectra in 31 bacterial species
07 September 2022
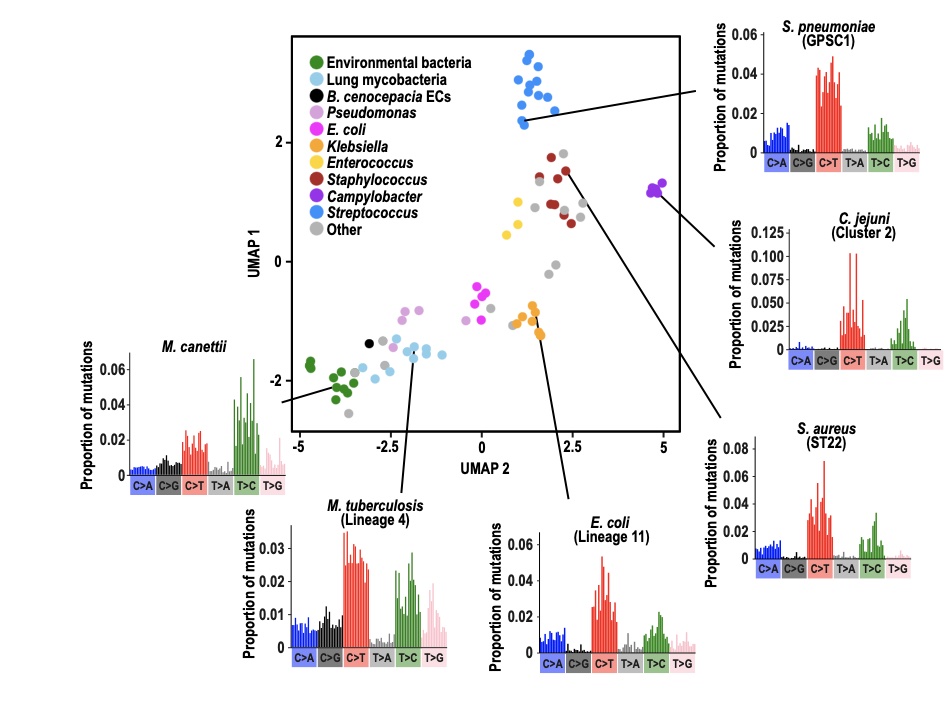
Led by Chris Ruis and a collaboration with Andres Floto in the Department of Medicine, our new paper entitled Mutational spectra analysis reveals bacterial niche and transmission dynamics is now on bioRxiv.
Mutational spectra (the patterns of base mutations and their surrounding nucleotide contexts) have been widely applied within the cancer field to infer the mutagenic drivers of cancers. This work has shown that individual mutagens drive specific mutational patterns. We reasoned that bacteria that live in different niches will be exposed to different sets of mutagens that will cause different mutational patterns. Therefore by charting the mutations acquired during evolution, we may be able to infer where a bacteria has lived and therefore the source(s) of infections. We tested this by calculating mutational spectra of 84 phylogenetic clades across 31 different bacterial species using a specifically developed pipeline, MutTui. This showed that mutational spectra are highly diverse across bacteria, both in base mutations and their contexts. However, we could observe groups of similar spectra that typically belonged to bacteria that are closely related and/or live in the same niche.
We examined the ability of mutational spectra to predict niche across several levels of comparison. We first showed that the mutational spectra of lung and environmental clades of Mycobacteria and Burkholderia cluster by niche. This is likely driven by elevated C>A mutations in the lung and higher T>C mutations in the environment. A decomposition analysis on the niche-specific mutations suggested that higher C>A in the lung is likely driven by tobacco smoke and reactive oxygen species, while elevated T>C is driven by environmental alkylating agents and nitro-PAHs. We next examined multiple niches within a single host and found that enteric and invasive lineages of Salmonella exhibit distinct spectra. Finally, we examined spectra of skin bacteria and identified a UV light-associated signature in the pan-skin bacterium Cutibacterium acnes that is not present in Staphylococcus epidermidis, which inhabits moist skin sites and is therefore hidden from the external environment.
Together, these results show that mutational spectra can predict general and specific bacterial niches and therefore reveal pathogen infection sites and transmission routes.